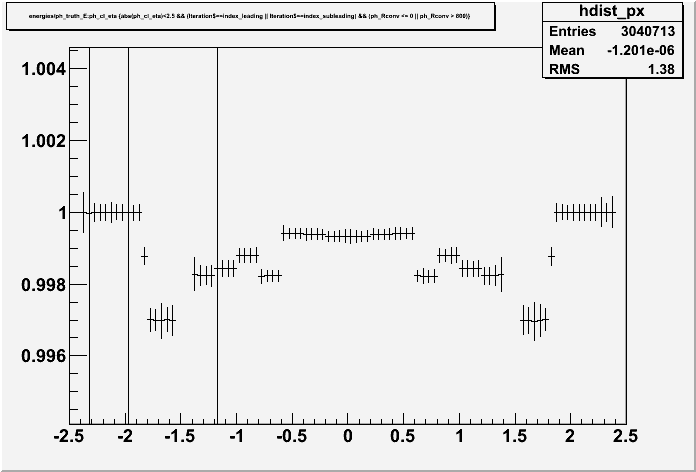
I am running two similar scripts, but I get completely different result:
import ROOT
dir_sys = "PSUp"
dir_nom = "all_mc"
fnom = ROOT.TFile(dir_nom + "/data-NTUP_output/mc12_8TeV.160009.PowhegPythia8_AU2CT10_ggH125_gamgam.merge.NTUP_PHOTON.e1189_s1771_s1741_r4829_r4540_p1344.root")
fsys = ROOT.TFile(dir_sys + "/data-NTUP_output/mc12_8TeV.160009.PowhegPythia8_AU2CT10_ggH125_gamgam.merge.NTUP_PHOTON.e1189_s1771_s1741_r4829_r4540_p1344.root")
tree_nom = fnom.Get("photon")
profile_nom = ROOT.TProfile("profile_nom", "profile_nom", 100, -2.5, 2.5)
tree_nom.Draw("energies/ph_truth_E:ph_cl_eta>>profile_nom", "(ph_Rconv >0 && ph_Rconv < 800) && (Iteration$==index_leading || Iteration$==index_subleading)", "prof")
histo_nom = profile_nom.ProjectionX()
histo_nom.SetDirectory(0)
tree_sys = fsys.Get("photon")
profile_sys = ROOT.TProfile("profile_sys", "profile_sys", 100, -2.5, 2.5)
tree_sys.Draw("energies/ph_truth_E:ph_cl_eta>>profile_sys", "(ph_Rconv >0 && ph_Rconv < 800) && (Iteration$==index_leading || Iteration$==index_subleading)", "prof")
histo_sys = profile_sys.ProjectionX()
histo_sys.SetDirectory(0)
ratio = histo_sys.Clone()
ratio.Divide(histo_nom)
canvas = ROOT.TCanvas()
ratio.Draw()
canvas.SaveAs("bo.png")root [4] TFile fnom("all_mc/data-NTUP_output/mc12_8TeV.160009.PowhegPythia8_AU2CT10_ggH125_gamgam.merge.NTUP_PHOTON.e1189_s1771_s1741_r4829_r4540_p1344.root")
root [5] photon->Draw("energies/ph_truth_E:ph_cl_eta>>hnom(100,-2.5,2.5)", "abs(ph_cl_eta)<2.5 && (Iteration$==index_leading || Iteration$==index_subleading) && (ph_R$
(Long64_t)1054554
root [6] hnom->SetDirectory(0)
root [9] TH1D* hhnom = hnom->ProjectionX()
root [10] hhnom->SetDirectory(0)
root [11] TFile fPSUp("PSUp/data-NTUP_output/mc12_8TeV.160009.PowhegPythia8_AU2CT10_ggH125_gamgam.merge.NTUP_PHOTON.e1189_s1771_s1741_r4829_r4540_p1344.root")
root [12] photon->Draw("energies/ph_truth_E:ph_cl_eta>>hdist(100,-2.5,2.5)", "(ph_Rconv >0 && ph_Rconv < 800) && (Iteration$==index_leading || Iteration$==index_subleading)", "prof")
Info in <TCanvas::MakeDefCanvas>: created default TCanvas with name c1
(Long64_t)1054610
root [13] TH1D* hhdist = hdist->ProjectionX()
hhdist->Sroot [14] hhdist->SetDirectory(0)
root [15] ratio = (*hhdist) / (*hhnom)
(class TH1D)256361552
root [16] ratio->Draw()I am getting completely different result: