I should have posted earlier, but didn’t know there was a “my ROOT app” category… anyway: I developed a python framework for the analysis of HEP data based on RDataFrame, called bamboo (see also the repository and documentation).
It mostly targets flat trees (where the branches contain simple numeric types and arrays of those) like CMS NanoAOD (the CMS open data format is very similar), and works as follows: based on the branches that are found (only the structure needs to be specified, not the full list of branches, so most differences between versions and data/MC do not need anything special), simple python classes are made that represent a lepton, a jet, a container of them etc. From these, expressions (mappings from an entry to a number or array) can be derived in compact python code, using a small set of helper methods, mostly for working with containers inside the same entry (event), and these can be used to define selection requirements and variables to plot (internally these are converted to C++ helper methods, which are declared with the cling interpreter, and RDataFrame nodes).
As an example, a plot with the sum of pT of jets with pT > 30 GeV that are not within 0.4 in ΔR of any electron or muon with pT > 10 GeV, for events with at least two muons with pT > 10 GeV, can be made as follows (see e.g. the implementation of the IRIS-HEP ADL benchmarks for a few more examples):
from bamboo import treefunctions as op
el10 = op.select(tree.Electron, lambda el : el.pt > 10.)
mu10 = op.select(tree.Muon , lambda mu : mu.pt > 10.)
hasTwoMuons = noSel.refine("hasTwoMuons", cut=(op.rng_len(mu10) >= 2))
cleanedJets30 = op.select(tree.Jet, lambda j : op.AND(
j.pt > 30.,
op.NOT(op.rng_any(el10, lambda el : op.deltaR(j.p4, el.p4) < 0.4 )),
op.NOT(op.rng_any(mu10, lambda mu : op.deltaR(j.p4, mu.p4) < 0.4 ))
))
plots.append(Plot.make1D("sumCleanedJetPt",
op.rng_sum(cleanedJets30, lambda j : j.pt), hasTwoMuons,
EqBin(100, 15., 200.), title="Sum p_{T} (GeV/c)"))
In the most common use case of filling a bunch of histograms for different selection stages, the user defines a python class that implements a definePlots method which returns a list of plots, and the input files are only processed once afterwards, when the histograms are retrieved. For the different selections there is a Selection object that keeps track of a set of cuts and weight factors, and is constructed by adding those to another selection (noSel above is the root of that hierarchy) - this maps to a Filter node (sometimes with a Define for the weight).
Having this python layer between the user code and RDataFrame also allows for some optimisation (expensive expressions are Defined and reused) and more advanced features: if configured to take jet systematic variations into account (with a bit of configuration code before), the fragment above will produce not one but 1+N histograms, and keep track of the associated RDataFrame graph branches (e.g. when plotting leading jet pT for events with two jets with pT > 30, there will be a Filter and attached Histo1D node for each variation). Since any C++ code can be called, it was also possible to reuse code to read a set of weights from a JSON file, and to calculate the jet variation on the fly (reusing the classes from CMSSW to read the corrections etc.).
The main benefit for the user is that the code to be written is a fairly compact representation of the actual analysis-specific choices (hence the link to analysis description languages), which helps to keep the overview, while the technicalities of turning that into reasonably efficient code are handled by bamboo and RDataFrame.
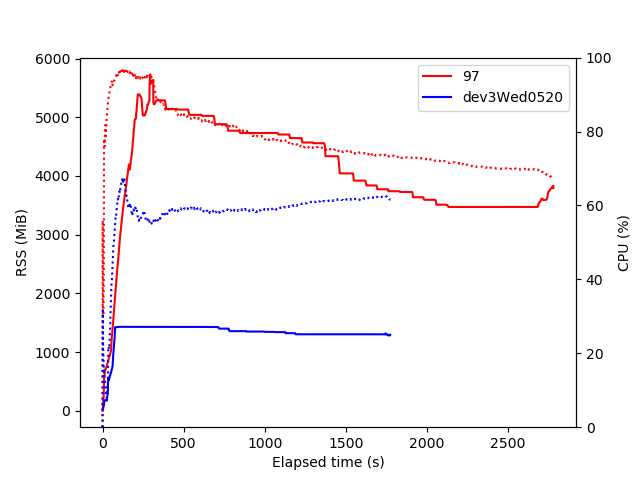
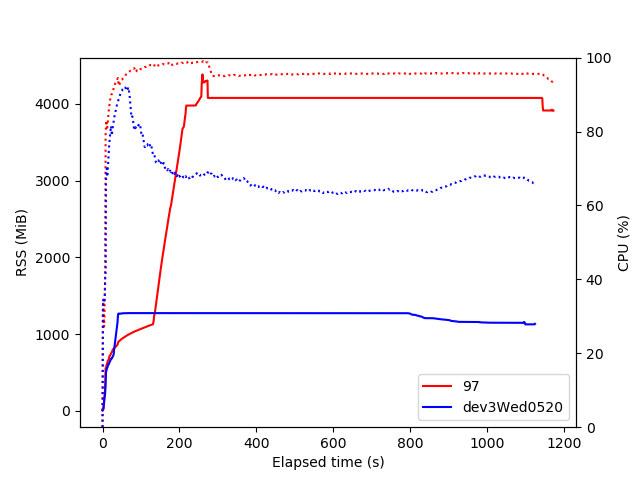
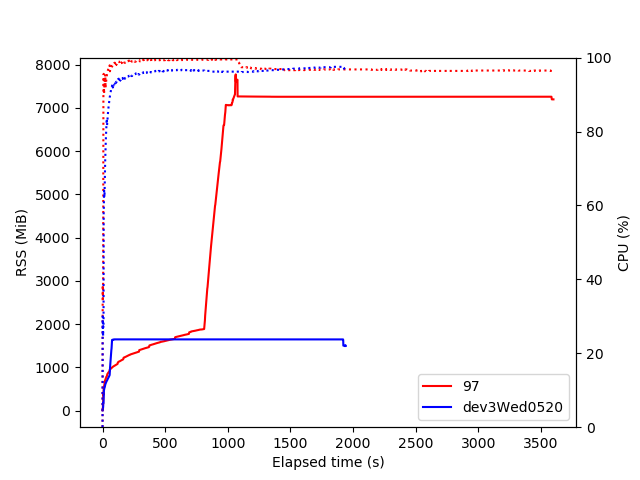
The main limitation for now is that we should not produce too many plots at once to keep memory usage under control (see RDataFrame+cling memory usage), but the performance and flexibility are sufficient for this to be used in several CMS analyses (five now, I think) - so many thanks to the ROOT team for the underlying framework. If I can help improve things by running some tests or benchmarks on these types of graphs I’ll be happy to do so.
Thanks,
Pieter